Defining a healthy microbiome and virome – a workshop on microbial functions
The Human Microbiome Action workshop on defining a healthy microbiome and virome was held on the 6th of October at the River Lee Hotel in Cork, Ireland. A total of 30 international top-ranked scientists in the microbiome field gathered in a hybrid format to discuss the current state of the art, as well as the future of microbiome science. In aiming to keep Europe at the forefront of microbiome research, the workshop activities were tailored to reach a consensus on research practices and study design needed to define a healthy microbiome and virome with future research. The focus was thereby to discuss a healthy functional core of the microbiome across different body sites, factors influencing its composition and function as well as a methodology to capture them.
The objective of the event was to gather experienced scientific leaders in the field to discuss the future of microbiome research. Initially, the focus was on definitions of health in humans and microbiomes, how and when to sample and what is the motivation for defining a healthy microbiome. This included gauging the current state of microbiome science, including methodologies, current data and findings, as well as missing links and gaps in knowledge. Progress was made on how to fill these gaps, discussing the study design and essentials for future microbiome studies. After each session, interactive software tools were used to collect input from the participants. These items will be analysed in the upcoming weeks and months and will lead to the primary outcome, a white paper with recommendations to shape the future microbiome research informing the European Union.
The workshop will have a consensus report as its main outcome, which will be presented to the EU, policymakers and relevant stakeholders in form of a white paper, comprising summarised recommendations on how future research should be conducted to define and monitor a healthy microbiome.
What is a healthy microbiome?
When debating how to define a healthy microbiome, the discussion revolved around whether it can either be defined as a function of the host’s health, as a function of microbiome characteristics, as a combination (the ‘holobiome’) or retaining the two elements independently. It was emphasised that microbiome health is a dynamic state space rather than a fixed characteristic. The group discussed whether different definitions are necessary depending on the research question asked.
The point of the appropriate use of language and terminology in the field was raised. As an example, the broad use of the term microbiome, where bacteriome would often be more appropriate, as the virome or mycobiome are often not included.
How do we measure it?
When discussing how to capture a healthy microbiome, experts agreed, based on the previous assumption that functionality is a key target for studying health, that shotgun metagenomics are likely preferred. Integration of long-read sequencing and other ‘omics approaches, such as metabolomics, is very promising for future research. Future study design could incorporate longitudinal elements, with sampling frequency increased at life-stages where microbiome composition is changing rapidly (e.g. infants and elderly).
When discussing factors influencing the microbiome, experts emphasised the importance of capturing these data using appropriate tools, instruments and methodologies and agreed that certain very important factors, such as gut transit time, remain under-captured in microbiome studies. It was also emphasised that there is less information on how such environmental factors shape core functionality than taxonomy.
The group also discussed the issue of stool samples being used as a proxy for the whole gastrointestinal tract. The experts suggest that specific priority should be given to research how representative stool samples are of the more proximal GI tract.
Lastly, the potential of linking microbiome studies with national health services where possible was highlighted.
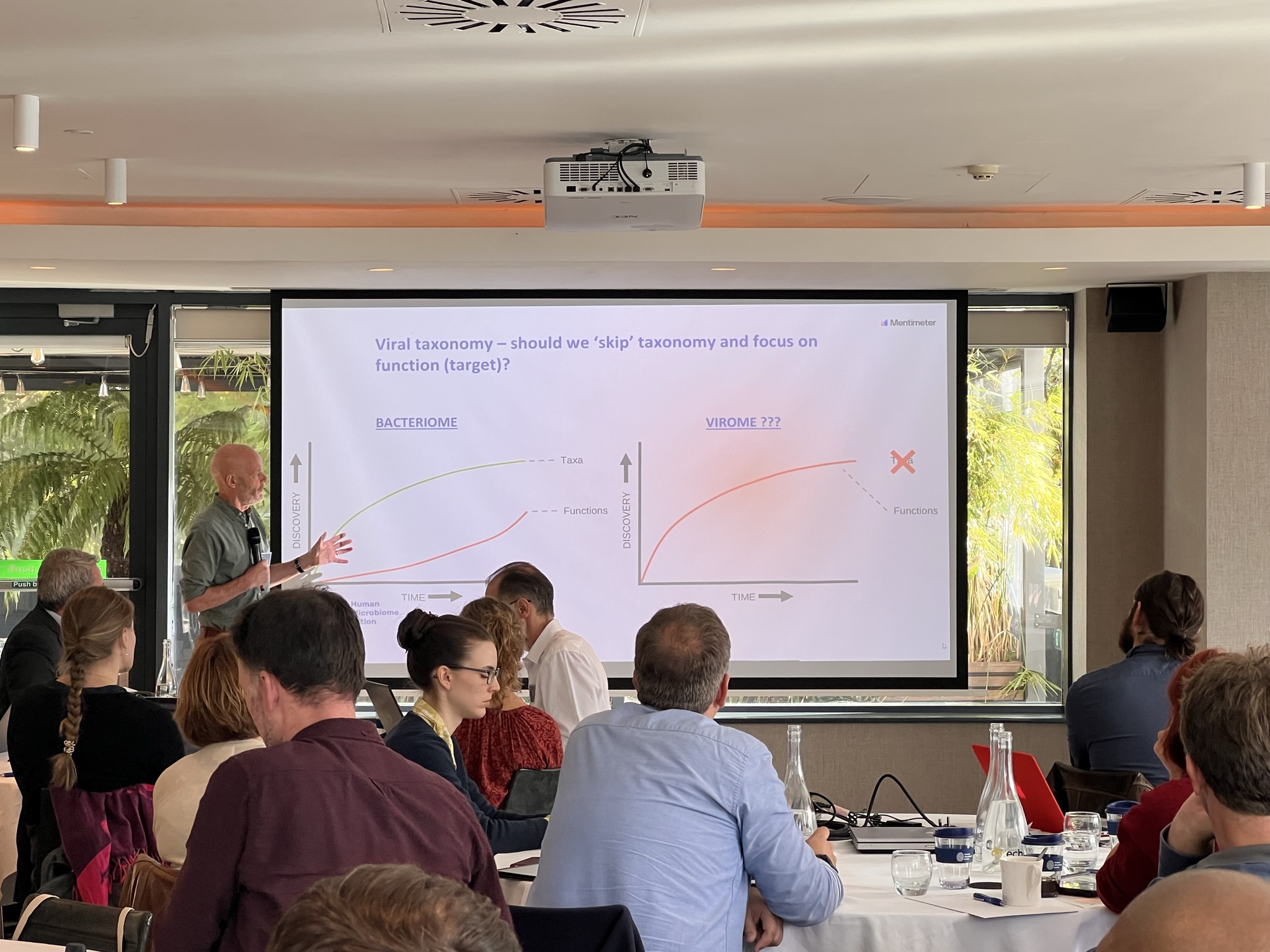
What is the role of the virome?
Gut virome researchers discussed within the larger group and in parallel, approaches to characterising the healthy virome. Points raised were challenges specific to virome research, such as sample processing and analysis and the trade-offs of different methodologies. Also, a discussion took place on the large degree of inter-personal variability, challenges with defining ‘functionality’ relevant to viruses, the significant degree of unexplored viral ‘dark matter’ and trying to link viruses with human health, given the complexity of interactions. There were also discussions on the spectrum of data globally, data gaps and the influence of environmental factors on the virome and how these could be addressed in future research.
The outcomes of this workshop will become available in form of a white paper at a later stage in the project.